VIRTUAL - Fourth BigBrain Workshop: Launch of the Helmholtz International BigBrain Analytics and Learning Laboratory (HIBALL) - a OHBM satellite event

Join us for the 4th BigBrain workshop and launch of the HIBALL on Friday June 26. Please check our programme below. We are proud to announce Fahmeed Hyder as keynote speaker. We will have two additional sessions with talks from senior and junior researchers from the BigBrain community, among others Timo Dickscheid, Sasha van Albada, and Blake Richards, followed by a series of lightning talks.
Please contact the program committee if you have any questions.
The Fourth BigBrain Workshop: Launch of the Helmholtz International BigBrain Analytics and Learning Laboratory (HIBALL) is a satellite event of the OHBM and is dedicated to the formal inauguration of HIBALL. The workshop will consist of invited talks covering recent work on the BigBrain data.
The newly established HIBALL (HIBALL), a collaboration between McGill University and Forschungszentrum Juelich aims to develop next-generation high-resolution human brain models with the help of state of the art Machine and Deep Learning (ML/DL) methods and high-performance computing (HPC) architectures. The lab especially targets the application of this future brain model for neuroimaging, brain-inspired AI research, and multi-scale brain simulation.
Abstract submission is closed.
Registration
Registration is now closed.
Please check your email for detailed login information.
The meeting will be streamed to YouTube here.
- Registration
- Programme
- Speakers
- Lightning Talks
- Call for Abstracts
- Partners
- Organizing Committee
- Contact
Registration
Registration is now closed.
Dial-information will be sent out by email the day before the meeting.
The meeting will be streamed to YouTube here.
Programme
| Time (EDT / UTC-4) | Time (CEST / UTC+2) | Description |
|---|---|---|
| 08:30 – 09:30am | 2:30 – 3:30pm |
Opening remarks and welcome notesOtmar Wiestler, President Helmholtz Association, Germany Sabine Sparwasser, German Ambassador to Canada Martha Crago, Vice-Principal (Research and Innovation), McGill University and Chair of Board of Directors HBHL Wolfgang Marquardt, CEO Forschungszentrum Jülich, Germany Remi Quirion, Chief Scientist of Québec Pawel Swieboda, Director General of HBP and CEO EBRAINS Katrin Amunts, Forschungszentrum Jülich, Germany Alan Evans, McGill University, Montreal, Canada |
| 09:40 – 10:20am | 3:40 – 4:20pm |
KeynoteFahmeed Hyder, Yale University, Magnetic Resonance Research Center Moderator: Alan Evans |
| 10:30 – 11:30am | 4:30 – 5:30pm |
Session 1Moderator: Katrin Amunts Timo Dickscheid, Forschungszentrum Jülich, Structural and functional organisation of the brain (INM-1) Sacha van Albada, Forschungszentrum Jülich, Computational and Systems Neuroscience (INM-6) Blake Richards, McGill University, Mila |
| 11:30 – 12:30pm | 5:30 – 6:30pm | lunch/dinner break |
| 12:30 – 1:45pm | 6:30 – 7:45pm |
Session 2Moderator: Timo Dickscheid Konrad Wagstyl, University College London Casey Paquola, McGill University, Montreal Neurological Institute and Hospital Jordan DeKraker, Western University Thomas Funck, Forschungszentrum Jülich, Structural and functional organisation of the brain (INM-1) Huifang Wang, Institut de Neurosciences des Systèmes, Marseille |
| 2:00 – 3:00pm | 8:00 – 9:00pm |
Session 3: Lightning TalksModerator: Boris Bernhardt Sherri Lee Jones, McGill University, Montreal Neurological Institute and Hospital Lindsay Lewis, McGill University, Montreal Neurological Institute and Hospital Jessica Royer, Casey Paquola, McGill University, Montreal Neurological Institute and Hospital Santanu Maiti, Forschungszentrum Jülich, Jülich Centre for Neutron Science (JCNS-1) Roxana Kooijmans, Markus Axer, Netherlands Institute for Neuroscience and Forschungszentrum Jülich, Structural and functional organisation of the brain (INM-1) Jorge Bosch-Bayard, McGill University, Montreal Neurological Institute and Hospital Pedro Valdes-Sosa, Cuban Neuroscience Center and The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China Jorge Riera Diaz, Department of Biomedical Engineering Kamil Uludag, University Health Network, Toronto Katja Heuer, Roberto Toro, Paris Institut Pasteur, Centre de Recherche Interdisciplinaire Mona Omidyeganeh, Claude Lepage, McGill University, Montreal Neurological Institute and Hospital Valerie Hayot-Sasson, Concordia University Chadi Barakat, Jülich Supercomputing Centre & University of Island Christian Schiffer, Forschungszentrum Jülich, Structural and functional organisation of the brain (INM-1) Andrea Brandstetter, Forschungszentrum Jülich, Structural and functional organisation of the brain (INM-1) Kai Kiwitz, C. and O. Vogt Institute of Brain Research, Düsseldorf |
| 3:15 - 3:30pm | 9:15 - 9:30pm | Last questions & Wrap up |
Speakers
Fahmeed Hyder: A 3D atlas of functional human brain energetic connectome based on neuropil distribution
Abstract:
Human cognition emerges from high energetic demands of the brain, but metabolism imposes limits on how much energy is allocated for functional and structural needs. Yet, how local variations in cellular and synaptic constituents of cortical networks affect regional variations in cerebral metabolism remains unknown. Based on principles that neuronal-glial glucose oxidation fuels signaling and nonsignaling activities of the neuropil, we created a high-resolution three-dimensional (3D) model of the human brain energetic connectome based on blueprints of cellular and synaptic densities. Explorations harnessing this 3D atlas revealed that globally distributed firing rates (~1.2Hz) dominated the heterogeneous metabolism, while cellular mass distribution accounted for metabolic homogeneity in the awake healthy human cortex. Transcortical gradients (i.e., from pial surface to white matter) of cellular/synaptic densities, cortical energetics, and neural firing revealed complex structure-function relationships. Stronger correlation between glucose oxidation and synapse density maps suggests that firing patterns are based on both local and distal connectivities rather than cellular mass distributions. Although synaptic density dictated spatial patterns of energetics and firing, nonsynaptic components accounted for a third of total energetics, implying that dysfunctions of enzymes involved in biosynthetic pathways can profoundly impact metabolism. The 3D atlas will permit energy-based constraint modeling of brain circuits across a range of spatial scales mapped by functional imaging.
Bio:
Dr. Hyder studies brain energy metabolism. He hypothesizes that mapping metabolic dysfunction (e.g., how much of energy reserves are used for cellular function vs. cell proliferation) can indicate early biomarkers of brain disorders and disease. To map brain’s physiology and chemistry with MRI and MRS advances and targeted delivery and monitoring of treatments, his laboratory conducts multidisciplinary functional and molecular imaging of neurodegeneration and neurooncology, requiring expertise in physics to chemistry to engineering to cancer biology to neuroscience. His contributions include development of high-resolution fMRI in preclinical models, calibrated fMRI for quantitative imaging of neuronal activity, and molecular imaging methods for pH, sodium, and temperature mapping and other physiological metabolic/hemodynamic parameters that are relevant in cancer diagnosis and treatment.
He received a bachelor degree in physical chemistry from Wabash College (in 1990) and a doctoral degree in biophysical chemistry from Yale University (in 1995). His work has produced nearly 200 peer-reviewed publications and invited presentations. He has written and edited books on functional imaging. He holds several magnetic resonance patents on molecular imaging. He has had continuous NIH support for over two decades and several NIH institutes have supported his research. He has renewed grants from different scientific funding agencies and he has received early career awards from various societies and agencies. He sits on editorial boards of several scientific journals and he reviews for many scientific journals spanning several disciplines. He serves on advisory panels of several funding agencies. He is a fellow of Sigma Xi the Scientific Research Honor Society, Academy of Radiology & Biomedical Imaging Research, and American Institute for Medical and Biological Engineering.
Timo Dickscheid: Towards automated mapping of cytoarchitectonic areas using Deep Learning
Abstract:
Mapping of cytoarchitectonic areas in the cerebral cortex is a key aspect of creating a multimodal human brain atlas. It segregates the brain into regions with a distinct spatial distribution of cell bodies, taking into account their shapes, sizes, and local density. The current de-facto standard for cytoarchitectonic mapping is a semiautomatic method, which identifies boundary definitions between two cortical areas in 1 micrometer resolution scans of stained tissue sections. While being reliable, precise, and observer-independent, the procedure requires close interaction with a human expert, and does not scale with steadily increasing quantity of histological sections from high-throughput microscopy. To this end, we evaluated deep convolutional neural networks (CNNs) for fully automated mapping of cytoarchitectonic cortical areas in ultrahigh resolution 2D scans. This is a challenging task for Deep Learning algorithms, due to the complex topology of the human brain, the inevitable variations in staining and tissue quality, different sectioning angles, and the very subtle properties of the cellular patterns that need to be uncovered. The talk will address the different Deep Learning strategies employed to achieve the first fully automatic mappings with limited training data, demonstrate practical impact in a real-world application with interactive training, and discuss existing limitations and future challenges. The presented methods have been used to compute highly detailed, full 3D maps of several cytoarchitectonic areas in the BigBrain that are now accessible online.
Bio:
Timo Dickscheid is heading the ‘Big Data Analytics’ group at the Institute of Neuroscience and Medicine (INM-1) at Forschungszentrum Jülich. He graduated in Computer Science at the University of Koblenz, and earned his PhD at the University of Bonn in 2011, where he worked on the 3D reconstruction of buildings from images. In 2010, he joined Forschungszentrum Jülich as a post-doc to build high-resolution 3D models of the human brain from microscopic images. After accepting a position as the head of Information Technology at the German Federal Institute of Hydrology in Koblenz in 2012, he returned back to Jülich in 2014 to build his own research group, which works on image analysis methods for building high-resolution models of the human brain, and software development for handling large microscopic image data. He regularly gives lectures in computer science at the Heinrich Heine University of Düsseldorf. In the Human Brain Project (HBP), Timo Dickscheid is the deputy leader of the neuroinformatics platform, and responsible for the development of a publicly accessible multi-level human brain atlas. He is also heading the research group on “AI methods for building ultrahigh resolution human brain models” of the local unit of Helmholtz AI, the German AI platform of the Helmholtz association.
Sacha van Albada: Large-scale spiking neural network models of resting-state dynamics in primate cortex
Abstract
How the complex connectivity structure of the cerebral cortex shapes its activity is a central question in neuroscience. To date, models of the cerebral cortex have seldom combined accounts of the dynamics with areas and across multiple areas. I will present a super-computational simulation study of resting-state activity in the cerebral cortex of the macaque which brings together microscopic and macroscopic descriptions of the structure and dynamics of the network. This work provides a stepping stone for developing large-scale dynamical models of human cortex, for which first efforts are underway.
Bio
Sacha van Albada is Junior Professor for Computational Neuroanatomy at the University of Cologne and group leader at Juelich Research Center in Germany. She studied Theoretical Physics at Utrecht University in the Netherlands, followed by a PhD in brain modeling at the University of Sydney. She is involved in the European Human Brain Project as deputy leader of the work package on cognitive architectures. She combines anatomical and physiological data from a wide range of sources to build neural network models of mammalian cerebral cortex. The aim is to understand relationships between cortical structure and dynamics, and to provide open-source models that serve as research platforms.
Blake Richards: Predictive cost functions in the neocortex
Abstract:
Neural computation is the product of two optimization procedures: natural selection and learning in the individual's lifetime. But, identifying what the neocortex has been optimized for, given its repeated circuit motifs and complex relationship to behaviour, is very challenging. Put another way, is it possible to say what cost functions have shaped the representations in the neocortex? Here, I will describe modelling data to show that the representations in the visual cortex of mice are likely shaped by a predictive cost functions.
Bio:
Blake Richards is an Assistant Professor in the School of Computer Science and the Montreal Neurological Institute-Hospital at McGill University and a Core Faculty Member at Mila. He was the 2019 Canadian Association for Neuroscience Young Investigator Award Recipient, and a CIFAR Canada AI Chairs announced. He is also a Fellow of the CIFAR Learning in Machines and Brains Program, and a Lab Scientist with the Creative Destruction Lab.
Konrad Wagstyl: BigBrain atlas of cortical layers: Linking cortical microstructure to in vivo measures of cortical structure.
Abstract:
The cerebral cortex exhibits a regionally varying microstructure which cannot readily be resolved using in vivo MRI. Nevertheless, cortical microstructure underpins many of the regional, developmental and pathological changes we can measure in vivo. In this presentation I will first discuss work that linked micro-anatomical laminar structure to MRI measures of cortical thickness. I will then present work done using the BigBrain, a 3D histological volume, to better characterise the microstructural origin of these MRI patterns of cortical thickness. We used an intensity profile-based approach to characterise how laminar structure varies between gyri and sulcal cortex. Using deep-learning, we extended this approach to segment 6 cortical layers in the BigBrain. We used these layers to confirm that the MRI patterns of cortical thickness were also present in the sensory cortices in a microscale histological dataset and to investigate which layers and neuronal subtypes drive these changes. In this way macroanatomical measures can be linked, via meso-scale layer thicknesses, to microscale neuron-level features.
Finally, in the process of creating tools for histological laminar analysis, we created open access tools to generate intracortical, equivolumetric surfaces. These surfaces follow contours of laminar structure across gyral and sulcal cortex, and can also be generated from in vivo MRI mesh surfaces.
Bio:
Konrad studied for his MBPhD at the University of Cambridge, studying for his PhD with Professor Paul Fletcher. During his PhD he spent a year working on the BigBrain at the MNI in the group of Prof Alan Evans. He is now a Sir Wellcome Research Fellow at the Wellcome Centre for Human Neuroimaging, UCL, using computational methods to find links between cortical microstructure and in vivo neuroimaging. As part of this work he co-leads the Multicentre Epilepsy Lesion Detection project, an open science collaboration to develop machine learning algorithms to automatically subtle focal cortical dysplasias in patients round the world.
Casey Paquola: Large-scale cortical gradients revealed by examination of intracortical microstructure
Abstract:
We recently developed an approach to model the similarity of microstructure profiles sampled in the direction of cortical columns using BigBrain. In doing so,we identified a system-level gradient of microstructural differentiation traversing from primary sensory to limbic regions, which followed shifts in laminar differentiation and cytoarchitectural complexity. Beyond conceptual insight, this also provided a crucial histological validation to develop the technique for in vivo magnetic resonance imaging (MRI). We found that large-scale gradients of microstructural differentiation in individuals, based on myelin-sensitive MRI, overlapped with core axes of cortical dynamics, allowing deeper insight into structure-function coupling at a macro-scale. In recent work, we have also demonstrated the utility of this approach in tracking intracortical microstructure throughout neurodevelopment and probing the complex neuroanatomy of the medial temporal lobe.
Bio:
Dr Casey Paquola is a postdoctoral fellow at the Montreal Neurological Institute-Hospital in the Multimodal Imaging and Connectome Analysis (MICA) lab of A/Prof Boris Bernhardt. She obtained her PhD in Medicine from the University of Sydney, Australia. Her research concerns the fundamental organisation and development of the cortex, with a special focus on combining histology and advanced neuroimaging to probe intracortical microstructure.
Jordan DeKraker: Data-driven parcellation and computational methods for generalizing results from the 3D BigBrain hippocampus to in-vivo MRI
Abstract:
The substructures of the hippocampus are challenging to map in detail in both histology and in in-vivo neuroimaging due to complex meso-scale folding that is difficult to appreciate using either method alone. MRI provides 3D context but is limited by resolution, while histology contains microscopic resolution but sampling is sparse and often limited to coronal sections through the hippocampal body. In this project we leveraged the fully reconstructed 3D histological sampling of the publicly available BigBrain dataset (Amunts et al., 2013) to generate a detailed model of the hippocampal subfields and numerous anterior-posterior folds. We demonstrate how this model can be used to inform hippocampal segmentation in MRI using a variety of methods, including 3D deformable registration, multi-planar resampling along the axis of hippocampal curvature, and topological or 2D surface-based registration. In particular, using a surface-based method which we previously developed to flatmap the hippocampus (DeKraker et al., 2018), we are able to show the inherent contiguity of each subfield and how this can be mapped to other hippocampi. Under this unfolding protocol, we then extracted a set of morphological and laminar measures of hippocampal tissue across its entire extent, and examined these features with respect to hippocampal subfields and anterior-posterior gradients proposed in the literature. Unsupervised clustering of these features revealed subdivisions that closely resemble gold standard manual subfield segmentations. Overall, our findings highlight new characteristics of internal hippocampal structure, and offer new avenues for its characterization with in-vivo neuroimaging. We argue that in this pursuit, a surface-based registration method provides the greatest flexibility to inter-individual differences in internal hippocampal folding.
Bio:
I am a Neuroscience Ph.D. student in the labs of Dr. Ali Khan and Dr. Stefan Köhler at the Robarts Research Institute and Brain and Mind Institute, University of Western Ontario. My thesis has focused on a variety of imaging and computational methods to better understand the structure of the human hippocampus, and in my Masters (UWO) and Bachelors (Carleton University) I have explored behavioural research in learning and memory in humans and rodents. I am currently developing a deep learning approach to hippocampal analysis in sub millimetric MRI, which I aim to soon release as a BIDSapp. I hope to apply these methods to large datasets and more deeply explore relations between meso-scale hippocampal structure, health, and theories of cognition as part of a postdoc. I am actively seeking collaboration or postdoc opportunities!
Thomas Funck: Towards an ultra-high resolution 3D neurotransmitter receptor atlas
Abstract:
Quantitative atlases of neurotransmitter receptor densities in healthy and pathologic human brains are important for characterizing normal and pathologic brain function and behaviour. Autoradiography can be used to visualize receptor density in 2D sections, but is challenging to reconstruct in 3D. In prior work, Karl Zilles and Nicola Palomero-Gallagher had acquired autoradiographs for 20 neurotransmitter binding sites from 3 donor brains. This data set has to date not been fully reconstructed into 3D because of the many unique challenges involved in performing such reconstruction for these data. This presentation describes the creation of a pipeline for producing the first ever set of ultra-high resolution (50μm) 3D atlases using this data. We illustrate its application on 4 different GABAergic neurotransmitter binding sites in the human brain. These 3D receptor atlases will provide a novel and unparalleled set of atlases for the neuroscience research community that will help better elucidate the functional anatomy of the human brain.
Bio:
Thomas has a PhD in Neuroscience from McGill University. During his PhD he studied receptor mapping using PET and autoradiography and is interested in creating receptor atlases of the brain using in vivo and in vitro data.
Huifang Wang: Workflow integrating The Virtual Brain (TVB) with data features of the Big Brain
Abstract
The inclusion of more physiological detail into models of brain function often passes unquestioned as a strategy towards the improvement of the model and its subsequent increased of predictive power. This approach is reminiscent of the situation in physics 150 years ago, when Maxwell wrote “... if we conceive of a being whose faculties are so sharpened that he can follow every molecule in its course, such a being, whose attributes are as essentially finite as our own, would be able to do what is impossible to us.” (Maxwell’s demon, 1872). Today we know that brute force addition of more detail and data finds quickly its limits of explanatory power and needs guidance from theory and adapted workflows. Here we propose a workflow integrating data features of the Big Brain into large-scale network models of The Virtual Brain (TVB) with the objective to increase the predictive power of brain models. This workflow is adapted from the already existing VEP workflow on epileptic patients, currently used in modified form in the clinical trial EPINOV. We will elaborate on the three workflow components which are important building blocks for the validation of multi-scale simulations: 1) identification of regionally variant functional data features from iEEG signals; 2) demonstration of identifiability of network symmetry breaking in human brain imaging signals; 3) demonstration of increased predictive capacity of TVB using anatomically correct regionally variant priors in a Bayesian model inversion framework.
Bio
Dr. Huifang Wang is a research engineer who works in the Institut de Neuroscience des Systemes (INS) with Dr. Viktor Jirsa in Marseille, France. Her current research interests focus on both theoretical parts: the personalized large-scale brain-region-specific models and their evaluations on the experimental datasets; and application parts: the personalized modeling and fitting for epileptic patient data. She obtained a PhD in Robotics in 2008 and did the first postdoc at the university of Pisa with Pr. Antonio Bicchi. Since 2012 she has worked in theoretical neuroscience at INS.
Lightning Talks
Sherri Lee Jones: An Atlas of the Human Hypothalamus at Ultra-High Resolution using the BigBrain
Content : Introduction. The hypothalamus is a sexually dimorphic brain structure recognized as a key region for contributing to sex differences in health and disease. Yet research using in vivo magnetic resonance imaging (MRI) of the human hypothalamus is scarce because of difficulties associated with discrimination of its nuclei and boundaries due to its small size, and because of intermingled gray and white matter within it. To circumvent this limitation, we used BigBrain, a digital reconstruction of the post-mortem brain of a 65 year-old healthy man (Amunts et al., 2013), which permits detailed observations of anatomical structures due to its ultra-high resolution and contrast, in order to create a 3D atlas of the hypothalamus.
Methods. Segmentation of the hypothalamus was performed on BigBrain (2015 release) using the Atelier 3D software (A3D) (Borgeat et al., 2007). The annotations were performed volumetrically on voxels at 20μm isotropic resolution. Identification of hypothalamic nuclei and boundaries were based on the Mai atlas (Mai et al., 2015) referring to the original histological publications. The “color gradients” feature in A3D facilitated visualization of hypothalamic substructures and boundaries. For each structure, the following workflow was used: 1) Manual segmentation was performed in the coronal plane every 2 to 20 sections depending on the size and shape of the structure. 2) Surfaces were automatically extracted, and verified for accuracy. 3) Contours were extracted from the mesh at a smaller spacing interval, then edited as needed. This process was repeated until each section was annotated. A closed surface was then obtained for each annotated structure. These surface were slightly smoothed locally based on normalized curvature operators in the normal direction (Desbrun et al., 2009; Kroon, 2019), while retaining their individual structure. This algorithm preserves the area on the surface and the volume inside the object and has a threshold to avoid moving too far from the demarcated points.
Results. Segmentation of the left and right hypothalamus, many of its nuclei as well as surrounding landmark structures were completed on BigBrain. These include preoptic, supraoptic, paraventricular, ventromedial and dorsomedial nuclei, as well as white matter tracts (anterior commissure, optic tract, fornix).
Conclusions. This is the first 3D atlas of the human hypothalamus at ultra-high resolution. The segmentation itself will be released as open-source. Future applications include 1) mapping it to MNI space and using it as a template in the analysis of an in-vivo MRI dataset to determine the influence of parental factors on the child’s neuroendocrine development; 2) using it as a validation tool for our novel in vivo manual segmentation protocol of the hypothalamus from high resolution structural MRI images (Jones et al. submitted). This atlas represents an important step in furthering our understanding of the human hypothalamus as a critical determinant in mental and physical health, especially with regard to sex differences.
References
Amunts K, Lepage C, Borgeat L, et al. (2013). BigBrain: an ultrahigh-resolution 3D human brain model. Science, 340(6139), 1472–1475. Borgeat L, Godin G, Massicotte P, Poirier G, Blais F, & Beraldin JA. (2007). Visualizing and analyzing the Mona Lisa. IEEE Computer Graphics and Applications, 27(6), 60–68.
Mai JK, Majtanik M, & Paxinos G. (2015). Atlas of the Human Brain. Academic Press.
Desbrun M, Meyer M, Schroder P & Barr AH. (1999). Implicit Faring of Irregular Meshes using Diffusion and Curvature Flow. Proceedings of the 26th annual conference on Computer Graphics and Interactive Techniques. 317-324.
Jones SL, Anastassiadis C, Dupuis M, Pruessner JC. A manual segmentation protocol of the hypothalamus and the pituitary gland using structural MRI. Submitted.
Kroon DJ. (2019). Smooth Triangulated Mesh (https://www.mathworks.com/matlabcentral/fileexchange/26710-smooth-triang...), MATLAB Central File Exchange.
Sherif, T., Kassis, N., Rousseau, M.-É., Adalat, R., & Evans, A. C. (2014). BrainBrowser: distributed, web-based neurological data visualization. Frontiers in Neuroinformatics, 8(149), 89. http://doi.org/10.3389/fninf.2014.00089
Primary authors : Dr. JONES, Sherri Lee (McGill University)
Co-authors : Dr. LEPAGE, Claude (McGill Centre for Integrative Neuroscience, McConnell Brain Imaging Center, Montreal Neurological Institute)
Dr. OMIDYEGANEH, Mona (Montreal Neurological Institute)
Dr. TOUSSAINT, Paule (McGill University, Montreal Neurological Institute and Hospital, McConnell Brain Imaging Centre)
Dr. LEWIS, Lindsay B. (McGill Centre for Integrative Neuroscience, McConnell Brain Imaging Center, Montreal Neurological Institute)
Dr. BORGEAT, Louis (National Research Council of Canada)
Dr. MASSICOTTE, Philippe (National Research Council of Canada)
Dr. ALTINKAYA, Ayça (McGill Centre for Integrative Neuroscience, McConnell Brain Imaging Center, Montreal Neurological Institute, Department of Biomedical Engineering, McGill University)
Dr. NGUYEN, Tuong-vi (Research Institute of the McGill University Health Center;Department of Psychiatry, McGill University)
Dr. SADIKOT, Abbas (McGill Centre for Integrative Neuroscience, McConnell Brain Imaging Center, Montreal Neurological Institute, Department of Biomedical Engineering, McGill University)
Prof. EVANS, Alan C. (McGill Centre for Integrative Neuroscience, McConnell Brain Imaging Center, Montreal Neurological Institute, Department of Biomedical Engineering, McGill University)
Prof. PRUESSNER, Jens C. (Department of Psychology, University of Konstanz)
Presenters : Dr. JONES, Sherri Lee (McGill University)
Lindsay Lewis: An updated MSM surface registration pipeline to bridge atlases across the BigBrain and FS / HCP worlds
Content : Reference brains in human brain mapping enable integration of multimodal data into a common framework (Evans et al. 2012). While MNI stereotaxic space has been widely adopted, reference cortical surfaces have been derived in differing ways from various pipelines. Commonly used population average surfaces include FreeSurfer's (FS) fsaverage (Fischl 2012), Human Connectome Project's (HCP) fs_LR (Van Essen et al. 2012), and the MNI152 (CIVET-generated: Lyttelton et al. 2007; see also Kim et al. 2005, Lepage et al. 2017). An additional reference surface of notable interest is that extracted from the 3D-reconstructed histological BigBrain model (Amunts et al. 2013, Lewis et al. 2014, Wagstyl et al. 2020). In the absence of a common framework across these structural pipelines, until recently, several high profile surface atlases in the FS / HCP world [e.g., HCP-MMP1.0 (Glasser et al. 2016), Brainnetome (Fan et al. 2016)] had neither been available for the MNI152 nor for BigBrain. Last year, we introduced the first surface registration pipeline linking these reference surfaces, as needed for a wide range of functional applications, but also for comparison and validation of the pipelines themselves (Lewis et al. 2019).
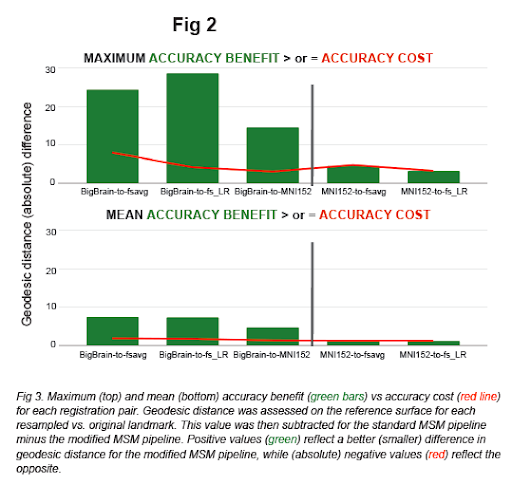
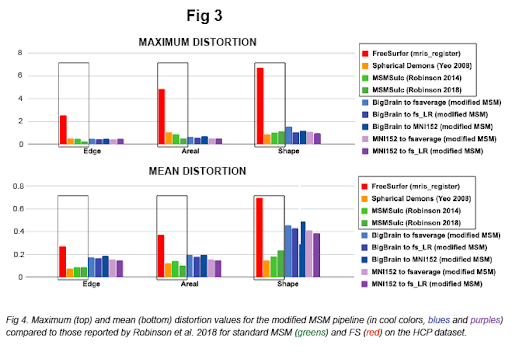
In the present work, we establish an improved pre-processing approach and implement a reparameterized multiscale pipeline via HCP's Multimodal Surface Matching (MSM) tool (Robinson et al. 2014, 2018) and HCP workbench (Marcus et al. 2011). Fig 1 demonstrates visualizations of the output resampled surfaces / atlases, Fig 2 shows higher accuracy (of our modified pipeline vs. standard MSM), and Fig 3 shows comparably low distortion values (as standard MSM).
Of particular interest, this work allows the high-resolution, histological BigBrain model to serve as an unprecedented cross-validation tool. Any surface atlases defined in fs_LR or fsaverage space may now be transposed to BigBrain space such that macroscopic parcellation boundaries derived from in vivo imaging can be directly compared to cytoarchitectural properties. Likewise, BigBrain's histological landmarks or cortical layers may be transposed to fs_LR and fsaverage.
Primary authors : Dr. LEWIS, Lindsay (MNI / McGill)
Co-authors : Dr. LEPAGE, Claude (MNI / McGill)
Dr. GLASSER, Matthew (Washington University in St. Louis)
Dr. COALSON, Timothy (Washington University in St. Louis)
Dr. VAN ESSEN, David (Washington University in St. Louis)
Dr. EVANS, Alan (MNI / McGill)
Presenters : Dr. LEWIS, Lindsay (MNI / McGill)
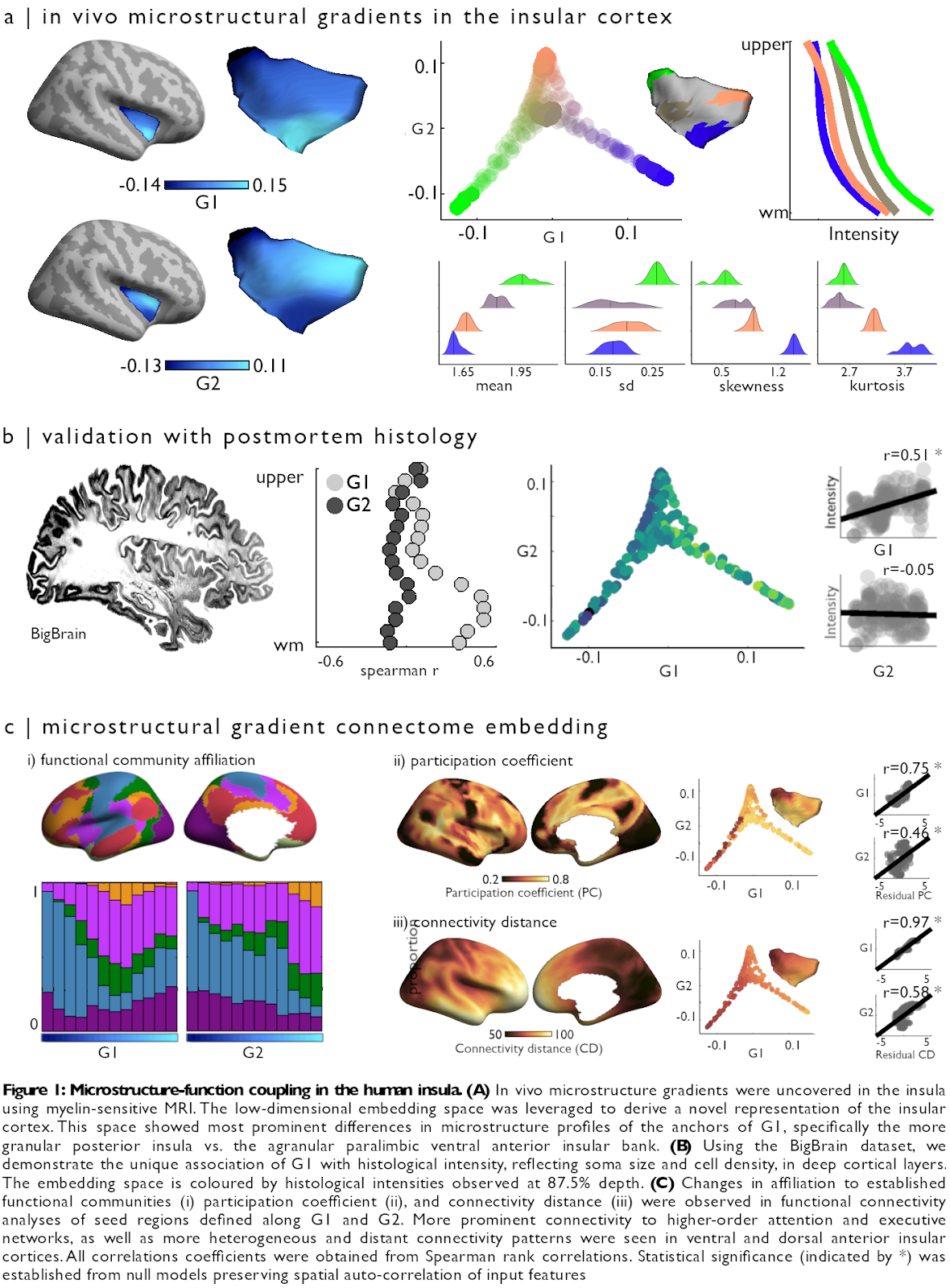
Jessica Royer, Casey Paquola: Myeloarchitecture gradients in the human insula: Histological underpinnings and association to intrinsic functional connectivity
Content : The insula is a core hub involved in multiple cognitive and socio-affective processes. Yet, the anatomical mechanisms that explain how it is involved in such a diverse array of functions remain incompletely understood. Here, we tested the hypothesis that changes in myeloarchitecture across the insular cortex explain how it can be involved in many different facets of cognitive function. Detailed intracortical profiling, performed across hundreds of insular locations on the basis of myelin-sensitive magnetic resonance imaging (MRI), was compressed into a lower-dimensional space uncovering principal axes of myeloarchitectonic variation. Leveraging two datasets with different high-resolution MRI contrasts, we obtained robust support for two principal dimensions of insular myeloarchitectonic differentiation in vivo, one running from ventral anterior to posterior banks and one radiating from dorsal anterior towards both ventral anterior and posterior subregions (Figure 1A). We then assessed the cytoarchitectonic underpinnings of each in vivo gradient, leveraging BigBrain – a 3D post mortem histological dataset. Analyses showed that the antero-posterior axis was mirrored in cytoarchitectural markers specifically in deeper cortical layers (r=0.51; p=0.01), and was robust to changes in sulco-gyral folding (partial r=0.44; p=0.05) (Figure 1B). Resting-state functional connectomics and ad hoc meta-analyses showed that myelin gradients in the insula relate to diverse affiliation to macroscale intrinsic functional systems, showing differential shifts in functional network embedding across each myelin-derived gradient. These gradients also followed shifts in the diversity of their functional connections, indexed by regional changes in participation coefficient (G1: r=0.69, p<0.01; G2: partial r=0.46, p<0.01), and the physical distance of functional connections (G1: r=0.95, p<0.01; G2: partial r=0.58, p<0.01) (Figure 1C). Collectively, our findings offer a novel approach to capture structure-function interactions of a key node of the limbic system, and suggest a multidimensional structural basis underlying the diverse functional roles of the insula.
Primary authors : ROYER, Jessica (Montreal Neurological Institute and Hospital, McGill University)
PAQUOLA, Casey (Montreal Neurological Institute and Hospital, McGill University)
Co-authors : LARIVIÈRE, Sara (Montreal Neurological Institute and Hospital, McGill University)
VOS DE WAEL, Reinder (Montreal Neurological Institute and Hospital, McGill University)
TAVAKOL, Shahin (Montreal Neurological Institute and Hospital, McGill University)
LOWE, Alexander (Montreal Neurological Institute and Hospital, McGill University)
BENKARIM, Oualid (Montreal Neurological Institute and Hospital, McGill University)
EVANS, Alan (Montreal Neurological Institute and Hospital, McGill University)
BZDOK, Danilo (Montreal Institute for Learning Algorithms (MILA); Department of Biomedical Engineering, Faculty of Medicine, McGill University)
SMALLWOOD, Jonathan (Department of Psychology, University of York)
FRAUSCHER, Birgit (Montreal Neurological Institute and Hospital, McGill University)
BERNHARDT, Boris (Montreal Neurological Institute and Hospital, McGill University)
Presenters : ROYER, Jessica (Montreal Neurological Institute and Hospital, McGill University)
Santanu Maiti: Quantifying nerve fibers/myelin in the Brain Section by Small Angle Scattering
Content : The structure and function of a brain are intricately linked to the structural connectome, i.e. the neurons and their connections [1]. The quantification of neural networks is important to understand brain function, dysfunction, and neurodegenerative disease. Here, we map a whole-brain section by small angle Neutron/X-ray scattering (SANS/SAXS) to investigate the microstructural insights of the brain tissue [2-4]. We quantify the spatial distribution of the nerve fibers and their degree of orientation. We also determine the orientation of myelinated axon and their orientational order over the section from the myelin Bragg peak. Finally, we compare them with 3D polarized light imaging (PLI) results to have a complete map of the connectome [5]. In the future, the scanning neutron/X-ray imaging can serve as an alternating technique for neuroimaging.
[1] C. S. von Bartheld et al., J. Comparative Neurology, 524, 3865 (2016). [2] H. Inouye et al., PLOS One. 9, e100592, (2014). [3] M. Georgiadis et al., NeuroImage, 204, 116214 (2020). [4] S. Foerster et al., Langmuir 31, 11678, (2015). [5] M. Axer et al., Front. Neuroinform., 5, 34 (2011).
Primary authors : Dr. MAITI, Santanu (Jülich Centre of Neutron Science (JCNS-1/IBI-8), Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich GmbH, Germany)
Co-authors : Dr. FRIELINGHAUS, Henrich (Jülich Centre for Neutron Science at Heinz Maier-Leibnitz Zentrum (JCNS-MLZ), Forschungszentrum Jülich GmbH, Germany)
Dr. DULLE, Martin (Jülich Centre of Neutron Science (JCNS-1/IBI-8), Forschungszentrum Jülich GmbH, Germany)
Mr. GRÄßEL, David (Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich GmbH, Germany)
Dr. AXER, Markus (Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich GmbH, Germany)
Prof. FÖRSTER, Stephan (Jülich Centre of Neutron Science (JCNS-1/IBI-8), Forschungszentrum Jülich GmbH, Germany)
Presenters : Dr. MAITI, Santanu (Jülich Centre of Neutron Science (JCNS-1/IBI-8), Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich GmbH, Germany)
Roxana Kooijmans, Markus Axer: Quantitative whole-brain multi-modal fibre and cell mapping in primates
Content : Detailed multi-modal architecture information is the basis for understanding function, dysfunction, and potential treatment of the brain. There are multiple efforts to generate complete and consistent maps for various species, but none addresses (quantitative) protein expression in combination with direct imaging of fibre distribution patterns. We have refined a method to integrate multi-channel, cell-type specific immunohistochemistry with polarized light imaging (3D-PLI), for revealing protein expression, as well as fibre architecture in 3D-space, in the same, full, primate brain sections. We image fibre tracts in 60μm-thick, unstained, brain sections at 1.3μm pixel size in-plane, using polarization microscopy (Axer et al.), in multiple species. Based on these measurements, regional fibre orientation maps are determined by means of big data analysis utilizing high- performance computing (JURECA supercomputer, JSC, Forschungszentrum Jülich), colour-coded and visualized. Subsequently, we unmount the tissue, and use immunohistochemistry to specifically label cells expressing calcium binding proteins parvalbumin, calbindin and calretinin, in the same section. Antibody- based staining in human tissue is typically less reliable than in mouse brain tissue, and the large size of full- hemisphere monkey or human brain section immunohistochemistry is methodically challenging. As a result, the combination of several antibodies in the same large-scale section is particularly novel. We remount the sections, and image them at 1μm resolution, using fast, full-colour, bright-field scanning. Subsequently, we segment the stained cell bodies using machine learning, and separate the different populations based on colour, to describe cellular distributions with high accuracy (Kooijmans et al.). This allows for acquiring more data at the same time, as well as relational data between targeted proteins. Finally, the two data sets can be aligned section-wise, using non-linear registration tools. This approach enables the visualization, segmentation, classification, and quantification of distinct cell populations, as defined by protein expression, in the context of the local and global fibre architecture, within the same section. Ultimately, we aim to obtain a complete, consistent, and multi-modal map of the brain, to be integrated into the atlas of the Human Brain Project, and made available to the international research community.
Primary authors : Dr. KOOIJMANS, Roxana (Netherlands Institute for Neuroscience) Dr. AXER, Markus (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich)
Co-authors : Mr. UPSCHULTE, Eric (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich)
Dr. DICKSCHEID, Timo (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich)
Prof. ZILLES, Karl (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich)
Prof. ROELFSEMA, Pieter (Netherlands Institute for Neuroscience)
Prof. AMUNTS, Katrin (Institute of Neuroscience and Medicine (INM-1), Research Centre Jülich)
Presenters : Dr. KOOIJMANS, Roxana (Netherlands Institute for Neuroscience)
Jorge Bosch-Bayard: From BigBrain to EEG/MEG source localization: alignment of neural generators with multimodal data
Content : We present the first preliminary efforts to extend the use of the Big Brain for electrophysiology. This is a joint work among researchers from the Cuban Neuroscience Center (CNeuro), the McGill Center for Integrative Neuroscience (MCIN) and the University of Electronic Science and Technology of China (UETSC). Inverse solutions are calculated to estimate the primary current distribution at the sources, using the Big Brain as the template. Two approaches were developed: the first one was a version of the Tomographic Quantitative EEG Analysis (qEEGt) at CNeuro. For this approach, a 3-spheres volumetric lead field was calculated over the Big Brain template co-registered to the MNI centroid, using the centroid segmentations for the skull and scalp. The Big Brain template was added to the CNeuro qEEGt toolbox, to obtain the source spectra over the Big Brain cortex, using the VARETA solution. Additionally, researches at UESTC calculated a realistic superficial lead field and applied the newly developed BC-VARETA inverse solution to estimate the primary current distributions at the sources in the time domain for a magnetoencephalographic (MEG) data. The MEG data was obtained from the Human Connectome Project (HCP) and the solution obtained for the Big Brain was compared to the solution obtained for the individual subject’s MRI. The detailed Big Brain segmentations, co-registered to MNI centroid were provided by researchers of the MCIN. These two works are the first attempts of using the Big Brain as a template for electrophysiology. The qEEGt toolbox is the first EEG facility added to the CBRAIN as a plugin for processing EEG information. It includes normative data freely released as open data for the first time. Future work will develop methods for using the Big Brain detailed anatomy to create more accurate forward models for the primary current estimation at the sources.
Primary authors : Dr. BOSCH-BAYARD, Jorge (Montreal Neurological Institute, McGill University, Montreal, Canada)
Co-authors : PAZ-LINARES, Deirel (Cuban Neuroscience Center and The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China)
AUBERT-VAZQUEZ, Eduardo (Cuban Neuroscience Center)
MARTINEZ-MONTES, Eduardo (Cuban Neuroscience Center)
GALAN-GARCIA, Lidice (Cuban Neuroscience Center)
WANG, Xindi (Montreal Neurological Institute, McGill University, Montreal)
Dr. LEWIS, Lindsay (Montreal Neurological Institute, McGill University, Montreal)
ARECES-GONZALEZ, Ariosky (Cuban Neuroscience Center and The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China)
CRESPO-BALTAR, Jose E. (Cuban Neuroscience Center)
PEREZ-RAMIREZ, Tania (Cuban Neuroscience Center)
LEPAGE, Claude (Montreal Neurological Institute, McGill University, Montreal)
Dr. TOUSSAINT, Paule (Montreal Neurological Institute, McGill University, Montreal)
EVANS, Alan (Montreal Neurological Institute, McGill University, Montreal)
VALDES-SOSA, Pedro (Cuban Neuroscience Center and The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China
Presenters : Dr. BOSCH-BAYARD, Jorge (Montreal Neurological Institute, McGill University, Montreal, Canada)
Pedro Valdes-Sosa: Precise electrophysiological connectivity with the Virtual BigBrain
Content : We describe a high-resolution estimator of brain EEG/MEG connectivity. Currently used estimators suffer from large amounts of “leakage”: spillover of estimated sources due to unacceptably high FWHM. This is a difficulty for their use with the high resolution BigBrain atlas as well as their use with TVB.
We define 3 families of Bayesian electrophysiological source imaging (ESI): activation (aESI), variability (vESI), and connectivity (cESI). We have shown ESI distortion and leakage are a consequence of miss-specifying prior distributions for aESI and subsequently using estimated sources time courses for vESI and cESI.
We show that a viable alternative is to target prior distributions directly on the source parameters to be estimated. For EEG/MEG source cross-spectra this can be achieved by a Hermitian graphical lasso prior—the HIGGS procedure. We show, with simulations and by means of a simultaneous EEG/ECoG monkey recording, that this allow substantial reduction of leakage.
We suggest future interfacing of BigBrain-based neural network models with multimodal neuroimaging data methods be based on symmetrical bayesian model fusion using the appropriate prior distributions.
Primary authors : Prof. VALDES-SOSA, Pedro (The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China and Cuban Neuroscience Center
Co-authors : PAZ-LINARES, Deirel (The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China and Cuban Neuroscience Center)
BOSCH-BAYARD, Jorge (Montreal Neurological Institute, McGill University, Montreal)
ARECES-GONZALEZ, Ariosky (The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China and Cuban Neuroscience Center)
EVANS, Alan (Montreal Neurological Institute, McGill University, Montreal)
Presenters : Prof. VALDES-SOSA, Pedro (The Clinical Hospital of Chengdu Brain Sciences Institute, University of Electronic Science and Technology of China UESTC, Chengdu, China and Cuban Neuroscience Center
Jorge Riera Diaz: EEG signatures of Ca2+ resonance in cortical pyramidal neurons—from monkeys to the Big Brain
Content : Ca2+ spikes initiated in the distal trunk of layer-5 pyramidal cells (PC) underlie nonlinear dynamic changes in the gain of cellular response, which is critical for top-down cognitive control. We present a minimal 2-compartment biophysical model that replicates the Ca2+ spike morphology and its critical frequency plus three other defining features of layer-5 PC synaptic integration: linear frequency-current relationships, backpropagation-activated Ca2+ spike firing, and a shift in the critical frequency by blocking Ih. Simulating 1,000 synchronized layer-5 PCs, we reproduced the current source density patterns evoked by Ca2+-spikes and reported its related scalp potentials on macaque monkeys. We discuss the utility of combining this minimal model and the detailed Big Brain cortical architecture to study the microcircuitry of agranular areas of the frontal lobe involved in cognitive control.
Primary authors : Prof. RIERA DIAZ, Jorge (Department of Biomedical Engineering, Florida International University)
Co-authors : Ms. HERRERA, Beatriz (Department of Biomedical Engineering, Florida International University)
Prof. SCHALL, Jeffrey D. (Department of Psychology, Vanderbilt University)
Dr. BOSCH-BAYARD, Jorge (McGill University, Montreal Neurological Institute and Hospital)
Presenters : Prof. RIERA DIAZ, Jorge (Department of Biomedical Engineering, Florida International University
Kamil Uludag: From BigBrain to fMRI: the role of hemodynamic modeling
Content : In this talk, I will outline a research line linking the BigBrain data to in vivo laminar fMRI in humans. High-resolution fMRI is increasingly used to non-invasively investigate mesoscopic functional circuits in the human brain, a domain previously only accessed with invasive methods in animals and intraoperatively in humans. Imaging on the mesoscopic level opens new avenues for human cognitive neuroscience: We cannot only image the patch of cortex involved in a specific brain function but distinguish feedforward from feedback processing streams within this patch based on their respective cortical depth activity profiles. Recently, we have introduced a new hemodynamic model of the laminar fMRI signal based on mass balance principles that accounts for vascular biases and distinguishes neuronal and vascular effects at the mesoscopic scale. In this talk, I will argue that regional specific microcircuit model derived from BigBrain data can be combined with the laminar fMRI model to interpret high-resolution fMRI data in humans in terms of excitatory and inhibitory neuronal activity.
Primary authors : ULUDAG, Kamil (University Health Network, Toronto)
Co-authors : HAVLICEK, Martin (Maastricht University)
Presenters : ULUDAG, Kamil (University Health Network, Toronto)
Katja Heuer, Roberto Toro: What is it like to be a Giant anteater? Cytoarchitectonic analysis of open data with MicroDraw.
Content : Introduction Brain histology provides unique information on the cellular structure of the brain. Histological data has been collected for more than a century, and is available for a wide variety of species and developmental stages. This type of data is, however, challenging to visualise, analyse and share. Here we present the current state of our Web tool MicroDraw, which aims at addressing these challenges through virtual distributed collaboration. We describe a complete analysis of the cytoarchitecture of a giant anteater brain, from data preprocessing to the representation of a quantitative cytoarchitectonic parcellation. Our results show that MicroDraw can be used to analyse high-resolution histological data, enabling distributed collaboration to improve our understanding of the cellular structure of the brain.
Methods
Histological slices of the brain of a giant anteater were obtained from the Michigan University collection website: https://brainmuseum.org/. The giant anteater is the largest of the 4 species of New World vermilinguans. Feeding on ants and termites, its geographic range extends from Central America to northern South America. The giant anteater is identified as vulnerable on the International Union for Conservation of Nature and Natural Resources Red List. The brain has been sectioned at 40 µm, 135 slices have been processed with Nissl staining, and made available in JPG format (image dimensions: 1152 x 864 pixels).
We downloaded the histological images, converted them to DZI format and uploaded them to MicroDraw (link to the data). Contours were drawn interactively using the threshold tool in MicroDraw plus manual editing. The contours were then downloaded using scripts written in Python 3 which query MicroDraw’s RESTful API.
To produce a globally coherent volume, the sections needed to be aligned. In a first step, the histological images were thresholded, downscaled to 25% in the coronal dimension, the result converted to a Nifti 1 volume and smoothed. The marching cubes algorithm was used for building a mesh, which was then manually corrected for topological errors. The smooth mesh was moved to histology space, and sliced to obtain aligned contours. Using the iterative closest point algorithm, the manually drawn contours were registered to the mesh contours. In a second step, a higher quality mesh was produced. The registered manual contours were shrunk to avoid self-collisions, transformed into images, and stacked into a nifti volume. The marching cubes algorithm was used to build an initial mesh, and its topology was then manually corrected. This higher quality mesh was transformed back into histology space, and used for the final registration of the manual contours. In a third step, we extracted grey level profiles. Starting from the manual contours, we generated inner contours by shrinkage. The original outer contours were then projected into the respective inner contour using an optimal transport algorithm. Based on these matched contour pairs, the histological images were sampled to obtain grey level profiles. The profiles were clustered using k-means to obtain a range of 2 to 30 clusters. Finally, we projected the cluster classes into the mesh.
Results
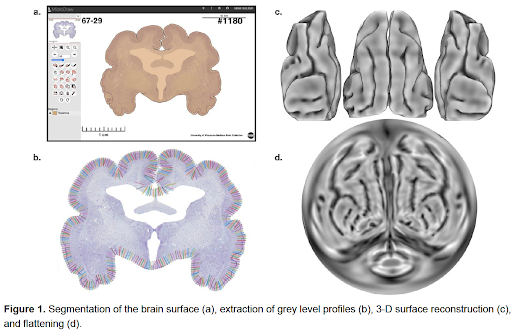
Our analysis combining MicroDraw and Python scripts enabled us to go from raw histological slice images to a quantitative cytoarchitectonic map based on grey level profiles. We obtained a coherently aligned volume of histological sections of the giant anteater brain. We created the contours of the pial surface for each slice using MicroDraw (Fig. 1a), and built a 3D surface reconstruction of the whole brain (Figure 1c, d). From our manual outer contours we obtained matching inner contours and sampled grey level profiles along the matched contours (Figure 1b). This approach allowed us to find clusters of grey level profiles and build a first map of the giant anteater brain based on cytoarchitectonic features (Figure 2).
Our grey level profile clusters recover cytoarchitectonic features. For the 2 clusters solution (Fig. 2a), for example, the clusters distinguish cortical from non-cortical areas such as the brainstem and the corpus callosum. The 5 clusters solution (Fig. 2b, c) provides an almost symmetric regionalisation of the brain, with a further distinction between gyral and sulcal cortices.
Conclusion
The current implementation of MicroDraw, in combination with our Python code, allows for a complete quantitative analysis of cytoarchitecture. This is the first time we implement a complete analysis workflow using MicroDraw and although there is still work to be done to ensure accuracy and reliability, our initial results are promising. MicroDraw is developed at Institut Pasteur in collaboration with Jülich Research Centre and the Montréal Neurological Institute. Its code is open and its development aims at following open science and software engineering best practices (ex., coding style, unit and end-to-end testing, continuous integration). MicroDraw is fast and lightweight, and can run on desktop computers and mobile devices. It could provide an open platform for data-sharing in our community, facilitating distributed scientific collaboration and citizen science.
Primary authors : Ms. HEUER, Katja (Max Planck Institute for Human Cognitive and Brain Sciences)
Dr. TORO, Roberto (Institut Pasteur)
Co-authors : DELETTRE, Céline (University Medical Center Eppendorf, Hamburg University)
GUI, Xiaoyun (Forschungszentrum Jülich)
Presenters : Ms. HEUER, Katja (Max Planck Institute for Human Cognitive and Brain Sciences)
Dr. TORO, Roberto (Institut Pasteur)
Mona Omidyeganeh, Claude Lepage: Non-linear registration of 1μm Histology Sections into 3D 20μm BigBrain Space
Content : Introduction: The manipulation of digitized histological sections of the human brain presents great challenges, due to the large size of the images, from processing to visualization. We present an algorithm to register 2D 1μm histological sections of the brain to the BigBrain, a 3D digital cytoarchitecture map of the post-mortem brain of a 65-year-old healthy male, reconstructed at 20μm [1]. Methods: The BigBrain was originally reconstructed from coronal sections, initially scanned at 10μm in-plane resolution. As new technology has become available, these sections have now been rescanned at 1μm in-plane resolution. Our goal was to map these data, now at cellular resolution, onto the BigBrain by co-registration of the sections at the two resolutions and then resampling of the 1μm sections to BigBrain 3D space using the existing transformations from the initial reconstruction of the volume. Data conversion: the scanned sections were first converted from big TIFF format to MINC (HDF-5 based with internal compression). Transformation Calculation: A global transformation was calculated for each 1μm section. This includes a concatenation of three individual transforms: 1) Non-linear transform from 1μm scanned to 10μm original (repaired), 2) Non-linear transform from original to MRI, and 3) Non-linear transform from MRI to aligned 3D BigBrain space. The latter two transforms originate from the initial reconstruction of the 3D BigBrain at 10μm. Blocking and Registration: The aligned 20μm image was divided into blocks to enable processing using standard tools without memory bottlenecks. The inverse transformation was applied to the corner points of each output block to estimate the extents (with padding) for cropping the input block of the non-registered 1μm section. The global transformation is applied to the cropped image to obtain the 1μm block in the aligned 3D BigBrain space. Reconstruction: The registered blocks were concatenated to form the registered 1μm section. Results: Our registration approach, by cutting the image into blocks, allows resampling of full histological sections of the human brain, at 1μm, whereas previously available tools could not be deployed on full sections due to prohibitive memory requirements. Each aligned, resampled section had a final size of 130,541 x 89,140 pixels for a file size of ~12GB (compressed). A typical block of size 13,100 x 9,000 pixels (10x10 blocks) took 8.4±2.0 minutes to transform, using less than 1.65GB memory. Block concatenation took ~65 minutes. Overall, the total processing time was ~15 hours on a single core, without memory bottlenecks, thus allowing simultaneous processing of multiple sections on multi-core compute nodes.
Primary authors : Dr. OMIDYEGANEH, Mona (McGill University, Montreal, Canada)
Dr. LEPAGE, Claude (McGill University, Montreal, Canada)
Co-authors : Dr. WAGSTYL, Konrad (University College London)
SPITZER, Hannah (Institute of Computational Biology, Helmholtz Zentrum München, Munich, Germany)
Dr. DICKSCHEID, Timo (Forschungszentrum Jülich)
Prof. AMUNTS, Katrin (Research Centre Jülich, Jülich, North-Rhine Westphalia)
Prof. EVANS, Alan C. (McGill University, Montreal, Canada)
Presenters : Dr. OMIDYEGANEH, Mona (McGill University, Montreal, Canada)
Valerie Hayot-Sasson: Sea: a hierarchical file system for efficient pipeline processing on HPC
Content : High Performance Computing (HPC) infrastructure is essential for the processing of ultra-high resolution datasets such as the BigBrain, due to the data’s computational and storage requirements. While such infrastructure provides ample resources for the processing of ultra-high resolution neuroimaging datasets, processing time can be significantly improved (up to 5x speedup) through incorporation data management tools (Hayot-Sasson et al., 2019). For instance, Big Data platforms leverage data management strategies such as data locality and in-memory computing to minimize data transfers and improve read and write bandwidth. Such strategies can be provided by distributed file systems, such as HDFS (Shvachko et al., 2010), or pipeline engines, such as MapReduce (Dean et al., 2004) and Apache Spark (Zaharia et al., 2016). HPC systems, however, generally favour the use of shared parallel file systems, such as Lustre (Braam et al., 2002), which maintain data and compute nodes separate, over the use of Big Data distributed file systems. Moreover, neuroimaging pipelines seldom use BigData frameworks potentially due to difficulties in implementing imaging pipelines using these frameworks (Mehta et al, 2017) and the need to refactor pre-existing pipelines for these frameworks.
To address the shortcomings of current Big Data frameworks for the efficient processing of large neuroimaging datasets, we propose Sea, a C++ library providing in-memory computing and data local performance to neuroimaging applications without any pipeline refactoring. Sea will require a hierarchy of file system paths, to determine where files should be accessed and stored, and a mount point with which the pipeline will use to interact with files managed by Sea. Whenever the application must access a file within the mount point, Sea will descend the hierarchy searching for the fastest available file system storing the file. If a file needs to be created, Sea will create this file in the fastest available file system provided. All files created by Sea will, by default, be flushed to persistent storage. Should the user not require some data to be written to persistent storage, the user will be able to specify this within a text file, further improving application performance.
This talk will present the architecture and implementation of Sea, and illustrate its value in the processing of BigBrain on HPC infrastructure.
Primary authors : HAYOT-SASSON, Valerie (Concordia University)
Co-authors : GLATARD, Tristan (Concordia University)
Presenters : HAYOT-SASSON, Valerie (Concordia University)
Chadi Barakat: Application Co-Design of a modular computing architecture for cellular BigBrain connecting the Canadian CBRAIN and German Supercomputing Infrastructures
Content : The development of parallel and scalable neuroscience application workflows across already established computing infrastructures face numerous challenges, because of the lack of common open standards and the different technologies used today. This talk will inform on the particular challenges of developing a joint architecture interconnecting the CBRAIN infrastructure [1] with the Juelich Supercomputing Cente (JSC) infrastructure based on modular supercomputing architecture (MSA) [2] principles. It is essential to the success of this joint infrastructure through its efficient use that its architecture is co-designed by collaborative neuroimaging and machine learning research applications.
This talk, therefore, first targets requirements obtained from neuroinformatics workflows that are part of the HIBALL (HIBALL) project [3]. While HIBALL requirements represent the priority, we are not losing sight of the broader landscape of infrastructure developments such as toolset activities in the Human Brain Project (HBP) or high-performance computing (HPC) resource provisioning through the FENIX infrastructure. The talk firstly addresses thus challenges and highlights requirements for enabling common computational workflows covering technical integration aspects of SLURM scheduler [4] and Boutique software containers [5] using JSON, and SnakeMake [6] approaches. We present initial ideas on how an overall architectural design looks and explain specific building blocks, such as how the JSON platform description already used in CBRAIN is one cornerstone of a joint architecture with JSC.
Since the volume and variety of neuroimaging datasets represent another challenge, the talk will also address the handling of imaging datasets for BigBrain analysis. That includes technical building blocks such as DataLad (based on git-annex) [7] and initial ideas to exploit hierarchical memory architectures for datasets in with HPC.
Bibliography
[1] Sherif T, Rioux P, Rousseau ME, Kassis N, Beck N, Adalat R, Das S, Glatard T, Evans AC. CBRAIN: a web-based, distributed computing platform for collaborative neuroimaging research. Frontiers in neuroinformatics. 2014 May 21;8:54.
[2] Suarez, E. ; Eicker, N. ; Lippert, T., Modular Supercomputing Architecture: from Idea to Production Contemporary High Performance Computing: From Petascale toward Exascale, Volume 3 FL, USA : CRC Press 3, : 3rd, 223-251 (2019), Online: http://hdl.handle.net/2128/22212
[3] Helmholtz HIBALL Project, Online: https://bigbrainproject.org/hiball.html
[4] Reuther A, Byun C, Arcand W, Bestor D, Bergeron B, Hubbell M, Jones M, Michaleas P, Prout A, Rosa A, Kepner J. Scheduler technologies in support of high performance data analysis. In2016 IEEE High Performance Extreme Computing Conference (HPEC) 2016 Sep 13 (pp. 1-6). IEEE, Online: https://doi.org/10.1109/HPEC.2016.7761604
[5] Glatard T, Kiar G, Aumentado-Armstrong T, Beck N, Bellec P, Bernard R, Bonnet A, Brown ST, Camarasu-Pop S, Cervenansky F, Das S. Boutiques: a flexible framework to integrate command-line applications in computing platforms. GigaScience. 2018 May;7(5):giy016, Online: https://doi.org/10.1093/gigascience/giy016
[6] Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012 Oct 1;28(19):2520-2, Online: https://doi.org/10.1093/bioinformatics/bty350
[7] DataLad, Online: https://www.datalad.org/
Primary authors : Mr. BARAKAT, Chadi (Forschungszentrum Jülich, Germany, and University of Iceland, Iceland)
Co-authors : Mr. EINARSSON, Pétur Helgi (Forschungszentrum Jülich, Germany, and University of Iceland, Iceland)
Prof. RIEDEL, Morris (Juelich Supercomputing Centre / University of Iceland)
Ms. HAYOT-SASSON, Valerie (Concordia University)
Prof. GLATARD, Tristan (Concordia University, Montreal)
Presenters : Mr. BARAKAT, Chadi (Forschungszentrum Jülich, Germany, and niversity of Iceland, Iceland)
Christian Schiffer: Convolutional Neural Networks for Efficient Mapping of Cytoarchitectonic Areas across Full Stacks of Histological Human Brain Sections
Content : Human brain atlases provide a reference system for mapping and localizing information from neuroscientific studies in different brains and under different modalities. Cytoarchitectonic mapping is considered the gold standard for defining the microstructural organization of the brain, as regional differences in the arrangement and composition of neuronal cells are an indicator of functional modules [1]. It requires to understand complex multiscale tissue patterns at microscopic resolution, including the presence of different types and sizes of neuronal cells as well as their spatial composition across larger spatial extents. The widely accepted semi-automatic observer independent method for cytoarchitectonic mapping of cortical areas[2] requires manual expert input for each tissue section, assumes approximately orthogonal sectioning angles, and is sensitive to significant histological artefacts. For being applied in a high-throughput scanning setup with full stacks of whole-brain sections, this approach is not practically feasible - a higher level of automization needs to be achieved, and a high robustness to arbitrary 3D orientations of the cutting plane as well as histological artefacts.
We present a complete and highly automatized workflow for mapping cytoarchitectonic areas in full stacks of histological sections of the same brain. It uses a Deep Learning model which is trained on a pair of expert annotations of the same brain area with considerable distance between the sections. The model learns to create all annotations in between with high accuracy, without assuming the stack to be 3D reconstructed or free of artefacts. This way it allows a human expert to label full stacks of sections efficiently. We present highly precise mappings of several cytoarchitectonic areas in the human brain, extending across thousands of consecutive histological sections, but based on less than 20 expert annotations, representing a manual annotation effort of a few days only.
[1] Amunts, Katrin, Este Armstrong, Aleksandar Malikovic, Lars Hömke, Hartmut Mohlberg, Axel Schleicher, and Karl Zilles (2007) “Gender-Specific Left-Right Asymmetries in Human Visual Cortex”. eng. In: The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 27.6, pp. 1356–1364.
[2] Schleicher, Axel, Katrin Amunts, Stefan Geyer, Patricia Morosan, and Karl Zilles (1999) “Observer- Independent Method for Microstructural Parcellation of Cerebral Cortex: A Quantitative Approach to Cytoarchitectonics”. In: NeuroImage 9.1, pp. 165–177.
Primary authors : Mr. SCHIFFER, Christian (Institute for Neuroscience and Medicine (INM-1), Research Centre Jülich)
Co-authors : Mrs. SPITZER, Hannah (Institute of Computational Biology, Helmholtz Zentrum München, Germany) Mr. KIWITZ, Kai (C. and O. Vogt Institute of Brain Research)
Mrs. UNGER, Nina (Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich, Germany)
Prof. HARMELING, Stefan (Institute of Computer Science, Heinrich-Heine University Düsseldorf, Germany)
Prof. AMUNTS, Katrin (Cécile and Oskar Vogt Institute of Brain Research, Univ. Hospital Düsseldorf, Heinrich-Heine University Düsseldorf, Germany)
Dr. DICKSCHEID, Timo (Forschungszentrum Jülich)
Presenters : Mr. SCHIFFER, Christian (Institute for Neuroscience and Medicine (INM-1), Research Centre Jülich)
Andrea Brandstetter: AI-supported cytoarchitectonic mapping and visualization of the lateral geniculate body in the BigBrain
Content : The lateral geniculate body is part of the metathalamus, and part of the visual pathway. It receives information from the retina of both eyes, and sends projections to the visual cortex. It comprises six layers, of which two (1, 2) are magno- and four (3, 4, 5, 6) are parvocellular. The six layers can also be subdivided into those receiving crossed fibers from the contralateral eye (1, 4, 6) and those receiving fibers from the ipsilateral eye (2, 3, 5). Thus, the lateral geniculate body shows a clear layer-wise architecture, but is not part of the cortex. In order to better understand the 3D-architecture of this important interface, we have mapped and 3D-reconstructed the lateral geniculate body using the BigBrain dataset.
The six layers were identified and manually delineated at every 30th section at cellular resolution in the BigBrain (distance between sections 0.6 mm). The Deep Learning model of the webtool ATLaS UI (by Christian Schiffer) was trained for recognizing cytoarchitectonic patterns based on manual delineations. In a second step, the model predicted delineations at sections in-between, i.e., on every section. Using this precise information of both the manual mapping and the AI-supported delineations we built a surface for each layer and visualized the results using the software ParaView. As a result, we have created an anatomical model that does not only show the overall shape of the lateral geniculate body, but also the fine-grained structure of its individual layers, and their relationship to each other.
Primary authors : Ms. BRANDSTETTER, Andrea (Institute for Neuroscience and Medicine (INM-1), Research Center Jülich)
Co-authors : Ms. BOLAKHRIF, Najoua (Institute for Neuroscience and Medicine (INM-1), Research Center Jülich)
Mr. SCHIFFER, Christian (Institute for Neuroscience and Medicine (INM-1), Research Centre Jülich)
Prof. AMUNTS, Katrin (Institute for Neuroscience and Medicine (INM-1), Research Center Jülich)
Presenters : Ms. BRANDSTETTER, Andrea (Institute for Neuroscience and Medicine (INM-1), Research Center Jülich)
Kai Kiwitz: Deep Learning Networks Reflect Cytoarchitectonic Features Used in Brain Mapping
Content : The distribution of neurons in the cortex (cytoarchitecture) differs between cortical areas and constitutes the basis for structural maps of the human brain. Deep-Learning approaches provide a promising alternative to overcome throughput limitations of currently used methods, but lack insight in how far they incorporate cytoarchitectonic features. We therefore analyzed the internal structure of a deep-learning based brain mapping tool for reflected cytoarchitectonic features and compared them to features of the currently used GLI profile approach. The tool consisted of a 10-block convolutional neural network and was trained to segment the primary and secondary visual cortex. Feature activations of the tool served to analyze resemblances to traditional cytoarchitectonic features and comparisons to the GLI profile approach. The analysis revealed resemblances to cellular, laminar- as well as cortical area related cytoarchitectonic features within the tool. It was able to develop feature activations that reflect the distinct cytoarchitecture of the segmented cortical areas with special regard to their laminar organization. The feature activations also compared well to statistical image criteria of the GLI profile approach. These results indicate an incorporation of relevant cytoarchitectonic features within the tool and mark it as a valid support in high-throughput mapping approaches.
Primary authors : Mr. KIWITZ, Kai (C. and O. Vogt Institute of Brain Research)
Co-authors : Mr. SCHIFFER, Christian (Institute for Neuroscience and Medicine (INM-1), Research Centre Jülich)
Prof. AMUNTS, Katrin (Forschungszentrum Jülich)
Dr. DICKSCHEID, Timo (Forschungszentrum Jülich)
Mrs. SPITZER, Hannah (Institute of Computational Biology, Helmholtz Zentrum München)
Presenters : Mr. KIWITZ, Kai (C. and O. Vogt Institute of Brain Research)
Call for Abstracts
Deadline extension! Please submit your abstract by June 9, 2020
Abstract submission opening date: February 4, 2020
Abstract submission deadline: June 5, 2020
You are welcome to submit short abstracts of current work and/or short proposals for future initiatives related to the BigBrain. Topics to be considered will include:
- Methods and algorithms to analyze the BigBrain, especially (but not necessarily) concerning
- Registration
- Segmentation and mapping of cortical layers, areas, and subcortical structures
- Visualization and annotation tools
- Deep learning and machine learning approaches
- HPC aspects of managing, storing and processing big data (high resolution and/or very large volume)
- Studies and teaching activities around the BigBrain
Please submit a short abstract (max. 1/2 page) by June 5, 2020. To submit an abstract you have to create an account. Submit a new abstract
Organizing Committee
Katrin Amunts, Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich
Alan Evans, The Neuro, McGill University
Deborah Rashcovsky, The Neuro, McGill University
Paule Toussaint, The Neuro, McGill University
Susanne Wenzel, Institute of Neuroscience and Medicine (INM-1), Forschungszentrum Jülich
Contact
Debbie Rashcovsky, Events Officer
Neuro Events, The Neuro, McGill University
Office: +1 514-398-6047 | Cellphone: +1 514-971-6047
Email: debbie.rashcovsky [at] mcgill.ca