Dissecting the gene regulatory function of monoubiquitylation on histone H2B:
Histone H2B is monoubiquitylated during the elongation phase of transcription by RNA polymerase II (RNAPII). This modification (termed H2Bub1) does not lead to H2B degradation by the proteasome, but instead seems to function as a regulatory signal. How it does this is not known, but the fact that H2Bub1 accompanies RNAPII transcription in all eukaryotic organisms argues that it plays a fundamental role in gene expression. Using the fission yeast model system (Schizosaccharomyces pombe) we have found an important connection between H2Bub1 and Cdk9. Unlike canonical CDKs, Cdk9 does not directly regulate the cell cycle but functions as a dedicated transcription factor whose activity drives RNAPII elongation on active genes. We have found that Cdk9 promotes H2Bub1 via a pathway involving two key molecular events: the phosphorylation of Spt5 and the consequent recruitment of Prf1/Rtf1. Both of these factors are conserved mediators of Cdk9 function in transcription elongation. Interestingly, H2Bub1 seems to negatively regulate an alternate Cdk9 pathway that involves the PAF complex and another phosphorylation target. Thus, our results place H2Bub1 at the heart of a key feedback regulatory mechanism controlling transcription elongation. Our current efforts focus on working out the molecular details of these pathways. This project is funded by the Canadian Institutes for Health Research.

P-TEFb/Cdk9 pathways regulating RNAPII transcription elongation. P-TEFb-dependent phosphorylation of Spt5 recruits Rtf1, a major co-factor for histone H2B ubiquitylation. Recruitment of the PAF complex involves additional P-TEFb substrates. These two pathways play distinct roles in regulation of RNAPII elongation.
Deciphering the “CTD code” for co-transcriptional histone modifications:
H2Bub1 is one part of a complex pattern of histone modifications that accompany RNAPII transcription of protein-coding genes. This pattern is a universal feature of transcription, yet how it is generated is still not known. The C-terminal domains (CTDs) of Spt5 and the large subunit of RNAPII itself (Rpb1) are important parts of these mechanisms. The Rpb1 and Spt5 CTDs are composed of multiple repeats of short amino acid motifs. The Rpb1 CTD contains the conserved heptad Tyr-Ser-Pro-Thr-Ser-Pro-Ser (numbered 1-7) while the Spt5 CTD is composed of a more variable motif that often begins with Thr. The CTDs are critical for assembly of the RNAPII elongation complex and interact with numerous factors, including histone modification enzymes. Remarkably, all of the phosphorylatable residues in these repeats are phosphorylated at different stages of transcription. This has led to the notion that patterns of phosphorylation along the CTD could constitute a “CTD code” that governs CTD-protein interactions. To begin to decipher how the CTD code dictates patterns of co-transcriptional histone modifications, we have used a systematic genetic approach in fission yeast involving mutants affecting CTD phosphorylation sites. We have also used chemical genetics to explore the functions of known CTD kinases (including Cdk9) in regulating histone modifications. Our data provide a starting point for detailed study of the molecular links between the RNAPII elongation complex and histone-modifying enzymes. This project is funded by the Natural Sciences and Engineering Research Council of Canada.
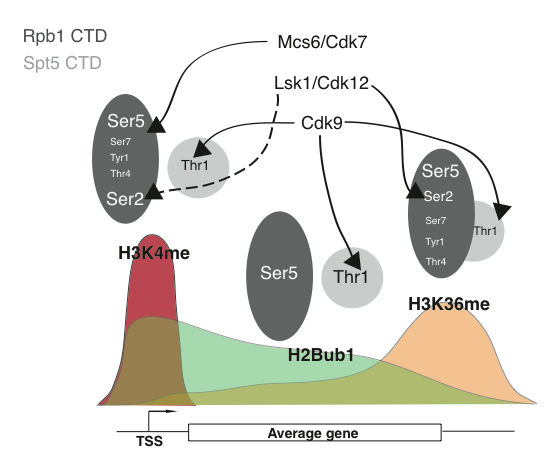
Patterning of co-transcriptional histone modifications by the Rpb1 and Spt5 CTDs. Arrows connect the transcription-associated CDKs to their targets on the Rpb1 or Spt5 CTDs above the three major co-transcriptional histone modifications (H3K4me-histone H3 lysine 4 methylation, H2Bub1-histone H2B ubiquitylation, H3K36me-histone H3 lysine 36 methylation). Solid arrows indicate positive regulation through the indicated phosphorylation event, whereas dotted arrows indicate
Targeting transcription elongation and histone modifications for treatment of cardiac hypertrophy:
We are beginning to translate our findings on the Cdk9-H2Bub1 pathway in fission yeast to mammalian cells. In collaboration with the lab of Terry Hébèrt (McGill Pharmacology), we are investigating the roles of Cdk9, its regulators, and downstream effectors (including histone modifications) on the hypertrophic response in cardiac myocytes. Cardiac hypertrophy (an enlargement of the heart muscle caused by increase in the size of individual myocytes) occurs in healthy individuals in response to exercise, but can also occur as a compensatory response in individuals with heart disease and atherosclerosis. This pathological form of cardiac hypertrophy turns out to be maladaptive and is associated with negative clinical outcomes. The activity of Cdk9 is required for the pathological hypertrophic response. We are interested in learning more about how Cdk9 (and potentially co-transcriptional histone modifications) drive gene expression changes associated with this response. Ultimately we hope to use these findings to develop new therapeutic strategies for treatment of heart disease. This project is funded by the Heart and Stroke Foundation of Canada.